Dna Replication
Before one cell can divide into two cells, the cell must make a copy of the cellular DNA so that after cell division, each cell will contain a complete complement of the genetic material. Replication is the cellular process by which DNA or the cellular genome is duplicated with almost perfect (and sometimes perfect) fidelity. The replicative process in prokaryotic cells, such as Escherichia coli ( E. coli ) cells, is best understood and will be described in detail, and the aspects that differ in replicating eukaryotic cells will be noted.
Replication starts by the separation of the strands of DNA and the formation of a local "bubble" at a specific DNA site called the origin of replication (ori). A helicase enzyme uses energy from ATP hydrolysis to effect this action. Single-strand DNA binding proteins stabilize the strands during the subsequent steps. The original DNA strands will function as the templates that will direct synthesis of the complementary strands. A nucleotide on the template strand will determine which deoxyribonucleotide (dNTP) will be incorporated in the newly synthesized strand. This replication
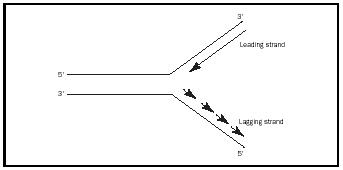
model is called semiconservative replication. The opening of the DNA produces two replication forks (see Figure 1), both of which will be the sites for replication. The forks will move in opposite directions relative to the replication machinery, with DNA replication occurring bidirectionally.
To initiate the synthesis of the two strands, a primer strand is needed, which is made by the enzyme primase. This small primer is an RNA molecule, which has a 5′ - and a 3′ -end . Replication requires the 3′ -hydroxyl group of the deoxyribose. This group is "attacked" by the phosphate of the incoming dNTP, because all DNA polymerases can extend DNA only from the 3′ -end (synthesis occurs in the 5′→3′ direction on the primer strand). Because the strands of DNA have opposite orientations, the replication process for each strand is considerably different. Extension of each strand requires very different operations that involve synthesis of a leading strand and a lagging strand. Leading strand synthesis, which progresses toward the fork in a continuous manner, begins with primase synthesizing a short RNA primer at the ori, followed by the action of DNA polymerase III, which incorporates deoxyribonucleotides into the strand, until strand synthesis is complete. If an incorrect nucleotide in incorporated, a proofreading activity in the mechanism removes it and the synthesis continues. Lagging strand synthesis progresses in the direction opposite to fork movement, with the new DNA strand synthesized in 1,000 to 2,000 nucleotide fragments called Okazaki fragments. Each fragment must also be initiated with a primer, followed by synthesis utilizing DNA polymerase III. When the Okazaki fragments are completed, DNA polymerase I both removes the RNA primers and simultaneously replaces the RNA with DNA. This occurs similarly for the single primer on the leading strand. The DNA fragments are then sealed together to produce a continuous strand by the enzyme DNA ligase.
The size of the genomic DNA in eukaryotic cells (such as the cells of yeast, plants, or mammals) is much larger (up to 10 +11 base pairs) than in E. coli (ca. 10 +6 base pairs). The rate of the eukaryotic replication fork movement is about fifty nucleotides per second, which is about ten times slower than in E. coli . To complete replication in the relatively short time periods observed, multiple origins of replication are used. In yeast cells, these multiple origins of replication are called autonomous replication sequences (ARSs). As with prokaryotic cells, eukaryotic cells have multiple DNA polymerases. DNA polymerase δ , complexed with a protein called proliferating cell nuclear antigen (PCNA), is thought to synthesize the leading strand, whereas DNA polymerase α is the replicase for the lagging strand. Eukaryotic genomes have linear DNA strands and require a special enzyme, called telomerase, to replicate the ends of the chromosomes.
SEE ALSO Base Pairing ; Deoxyribonucleic Acid (DNA) .
William M. Scovell
Bibliography
Garrett, R., and Grisham, Charles M. (1995). Biochemistry. Fort Worth: Saunders College.
Nelson, David L., and Cox, Michael M. (2000). Lehninger Principles of Biochemistry , 3rd edition. New York: Worth.
Comment about this article, ask questions, or add new information about this topic: